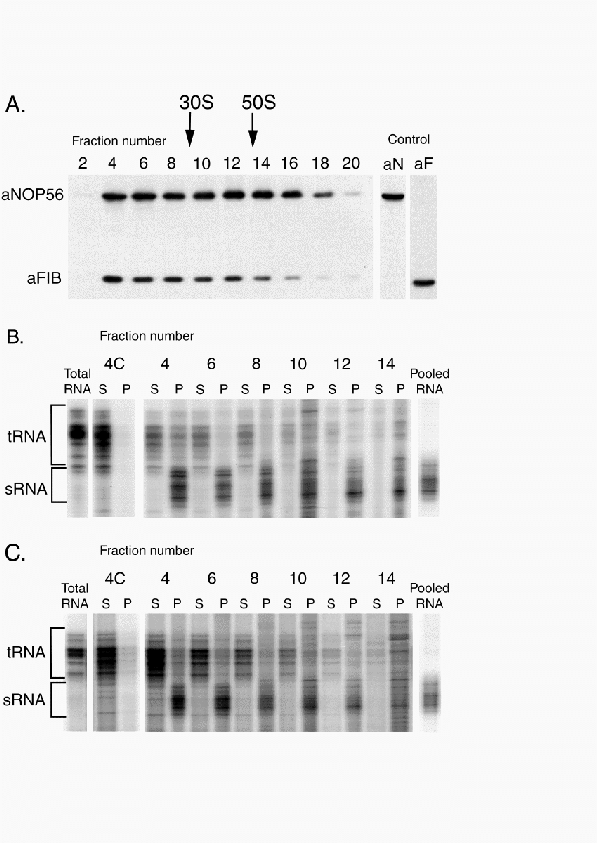
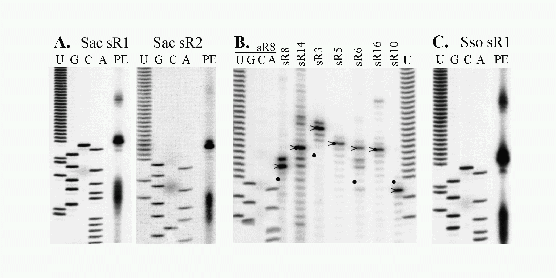
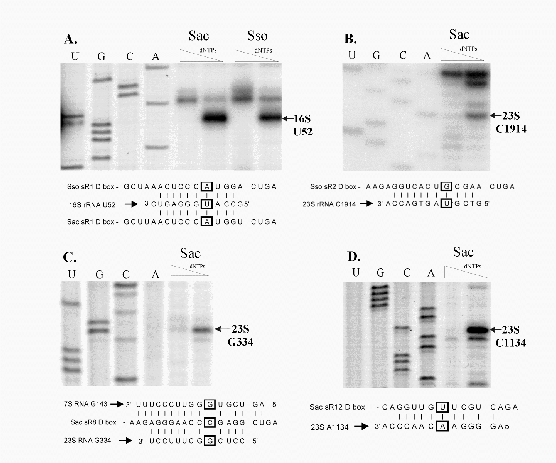
Figure 5.2:
Detection and 5' end mapping of sRNAs from S. acidocaldarius and S. solfataricus.
Primers specific for the D box guide region of Sac sR1 to sR17 were 5'
end labeled with
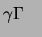 P-ATP and polynucleotide kinase and used in
extension reactions with total RNA (20
P-ATP and polynucleotide kinase and used in
extension reactions with total RNA (20 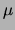 g) isolated from
S. acidocaldarius as template.
g) isolated from
S. acidocaldarius as template.
(A) The extension products obtained with Sac sR1 and sR2 specific
oligonucleotide primers were run alongside a DNA sequence ladder
generated with the same primers and Sac sR1 or sR2 cDNAs as
template.
(B) The extension products obtained with Sac sRNA-specific primers and
run with the DNA sequence ladder generated with Sac sR8 cDNA
clone. For each extension reaction, the approximate positions of the
5' terminal nucleotide in the corresponding cDNA is indicated by a
(>) beside the lane. [Note: there is an inconsistency in fragment
lengths for this experiment between the Dennis lab (gel picture here)
and my own primer extensions. This experiment will be repeated and
the conflict resolved before submission of this manuscript.]
(C) The primer extension reaction was as in (A) except that the primer
was specific to Sso sR1 and total RNA from S. solfataricus was used as
template. The DNA ladder was generated using Sac sR1 primer and the
Sac sR1 cDNA as template. The Sac and Sso primers differ at two
internal positions but have the same 5' and 3' end location.