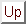


Next: Small Nucleolar RNAs in Genomes.2
Up: A Computational Screen for Yeast.2
Previous: Acknowledgments
All computer code, snoRNA search results, oligonucleotide primers,
rRNA primer extensions gel, and other referenced data can be found on
line [Lowe & Eddy, 1998]. All new snoRNAs (snR48,
snR50-snR71) have been submitted to the Saccharomyces cerevisiae
Genome Database (SGD; http://genome-www.stanford.edu/Saccharomyces/),
and can be accessed directly by searching for SNR locus names (e.g.,
``SNR50'', or ``SNR*''). Sequences are available in Genbank by accessions
AF06461-AF064283 for snR48, snR50-snR71, respectively. Other yeast
snoRNA Genbank accession numbers are as follows: snR190 and U14
(X96815), U18 (U12981), U24 (Z48760), snR13 (U16692), snR38 (U26012),
snR39 (U26011), snR39b (X94605), snR40 (U26015), snR41 (U26016), snR47
(U56648), Z2-Z8 (Z69294-Z69300), Z9 (Z70300).
Figure 4.1:
Schematic diagram of snoRNA search algorithm.
 |
Table 4.1:
Summary of states within snoRNA probabilistic model.
State numbers correspond to Figure 4.2. ``Ungapped
HMM'' states represent fixed-length conserved sequence motifs. The
state for the terminal stem is analogous, but models base pairs rather
than single positions (e.g., a stochastic context-free grammar, SCFG
[Durbin et al., 1998] instead of a hidden Markov model, HMM). Duration models for gaps
are estimated from binned length distributions (e.g., the probability
that a gap will be 11-20 nt, 21-30 nt, etc.). The guide state is a
hidden Markov model dependent on the rRNA target sequence; it includes
terms for the probability of starting the complementarity at a given
position relative to rRNA (this probability is high near known
methylation positions), the length of the complementarity, and the
probability of mismatches and noncanonical base pairs in the
complementarity. For each state, the most common feature (``consensus'')
is shown to indicate the overall pattern we search for. The best,
average, and worst feature scores are given for 41 methylation guide
snoRNAs as an indication of the relative contribution of each state to
the overall information in the model. For more detail, see the program
source code [Lowe & Eddy, 1998].
|
| State |
|
|
|
Feature Score (bits) |
| number |
[0pt]Feature |
[0pt]Model |
[0pt]Consensus |
Best |
Average |
Worst |
| 1 |
Terminal Stem |
SCFG, 4-8 bp |
6 bp (when present) |
7.60 |
3.09 |
0.35 |
| 2 |
Box C |
7 bp ungapped HMM |
AUGAUGA |
12.73 |
11.63 |
5.84 |
| 3 |
Gap |
Duration model |
Length 6-10 bp |
-1.59 |
-2.09 |
-4.76 |
| 4 |
Guide Sequence |
HMM |
12 bp duplex |
15.67 |
11.11 |
2.54 |
| 5 |
Box D' |
4 bp ungapped HMM |
CUGA |
7.34 |
4.85 |
-3.74 |
| 6 |
Gap |
Duration model |
Length 36-45 bp |
-1.59 |
-2.43 |
-5.36 |
| 7 |
Box D |
4 bp ungapped HMM |
CUGA |
8.05 |
7.92 |
5.43 |
| 8 |
Gap |
Duration model |
Length 56-75 bp |
-1.50 |
-2.10 |
-4.17 |
| 9 |
Guide Sequence |
HMM |
14 bp duplex |
18.96 |
13.98 |
9.95 |
|
Table 4.2:
C/D box snoRNAs in S. cerevisiae that function as methylation guides.
(see legend previous page)
|
|
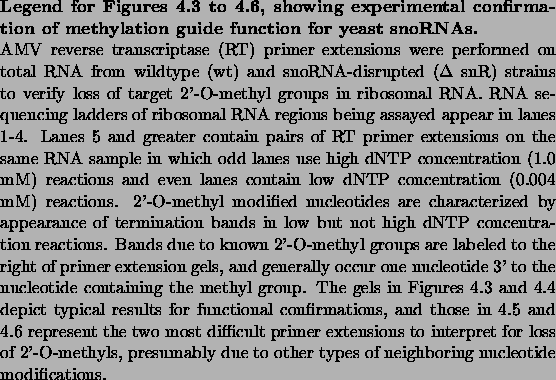
Figure 4.3:
Experimental confirmation of methylation guide function for
snoRNAs snR60, snR50, snR72, and snR40.
Loss of 2'-O-methyl bands in low dNTP-concentration
reactions for mutant strains (even lanes 8 and greater) relative to
the wildtype strain (lane 6) indicates loss of the methylation site
and thus functional confirmation. Polyacrylamide gel electrophoresis
of primer extensions using 32P end-labled primers annealing to 25S
rRNA from position 914-939.
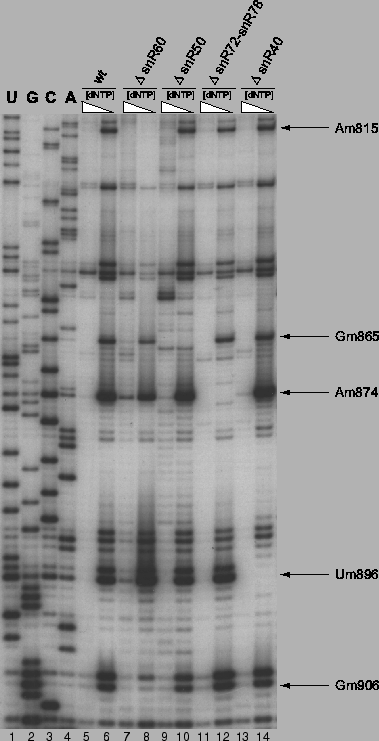 |
Figure 4.4:
Experimental confirmation of methylation guide function for
snoRNAs snR75, snR47, snR63, and snR13. Loss of 2'-O-methyl bands in
low dNTP-concentration reactions for mutant strains (even lanes 8 and
greater) relative to the wildtype strain (lane 6) indicates loss of
the methylation site and thus functional confirmation. Polyacrylamide
gel electrophoresis of primer extensions using 32P end-labled
primers annealing to 25S rRNA from position 2305-2328.
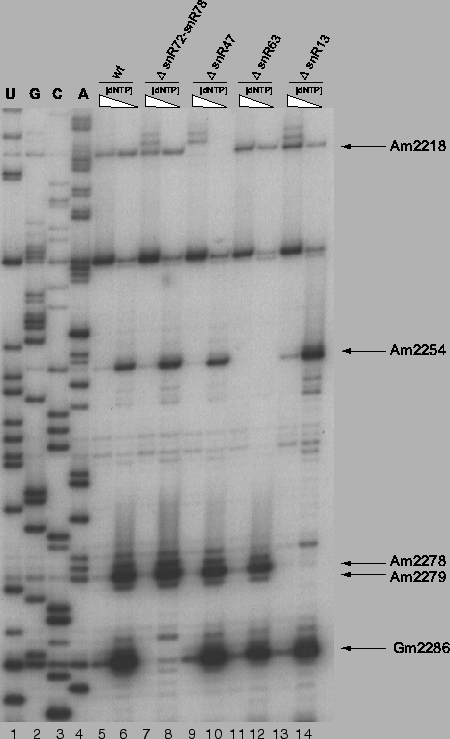 |
Figure 4.5:
Experimental confirmation of methylation guide function for
snoRNAs snR40 and snR55
Loss of 2'-O-methyl band in low dNTP-concentration
reaction for mutant strain (lanes 8 & 10) relative to the wildtype strain
(lane 6) indicates loss of the methylation site and thus functional
confirmation. Polyacrylamide gel electrophoresis of primer extensions
using 32P end-labled primers annealing to 18S rRNA from position
1291-1315.
 |
Figure 4.6:
Experimental confirmation of methylation guide function for
snoRNA snR70. Loss of 2'-O-methyl bands in low dNTP-concentration
reactions for mutant strains (lane 8) relative to the wildtype strain
(lane 6) indicates loss of the methylation site and thus functional
confirmation. Polyacrylamide gel electrophoresis of primer extensions
using 32P end-labled primers annealing to 18S rRNA from position
1652-1675.
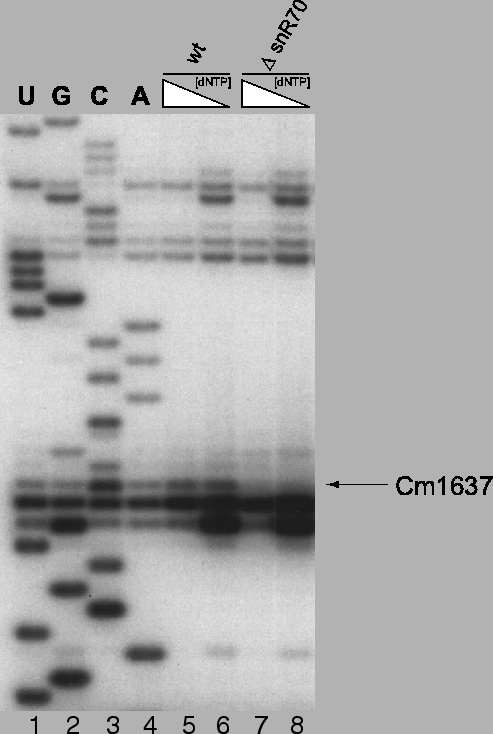 |
Figure 4.7:
snoRNA primer extensions demonstrating expression of newly
identified methylation guide snoRNAs.
Reverse transcriptase primer extensions on total RNA from wildtype and
snoRNA-disrupted strains. 32P end-labeled primers complementary to
internal snoRNA sequence for snR54 (lanes 1a,b), snR51 (lanes 2a,b),
snR63 (lanes 3a,b), snR55 (lanes 4a,b), snR68 (lanes 5a,b), snR70
(lanes 6a,b), snR71 (lanes 7a,b) were used. snoRNA expression in
wildtype RNA reactions (lanes 1a, 2a,...7a) was confirmed, as was
loss of expression in snoRNA-deleted strains (lanes 1b, 2b,...7b).
The snR54 internal snoRNA primer was included in all reactions as a
positive control of intact RNA and active primer extension. RNA
sequencing ladders of unrelated sequence are included on either side
of snoRNA primer extensions for fragment size reference.
|
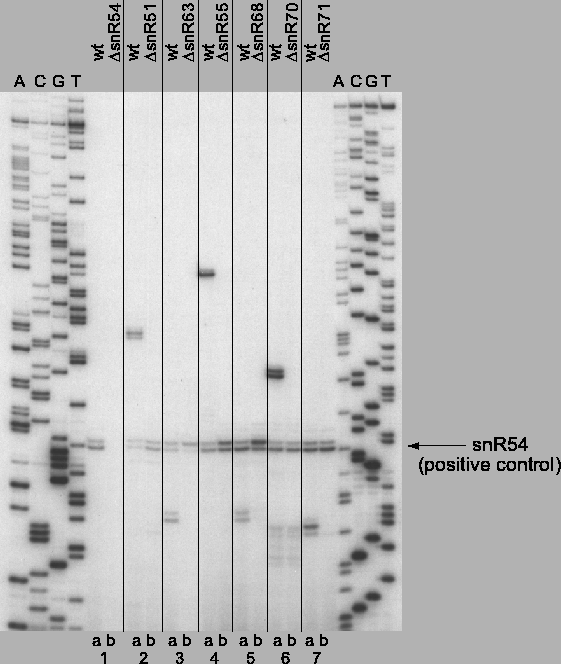 |
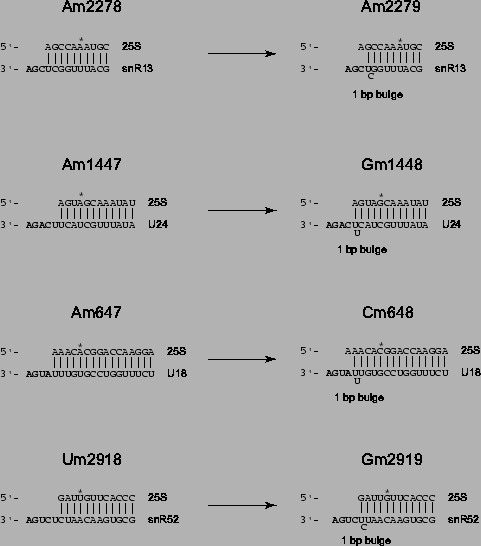
Figure 4.8:
Model for Addition of Adjacent 2'-O-methyls via the same
snoRNA. Listed are the four instances in yeast rRNA in which
2'-O-methyl groups occur just one nucleotide from other 2'-O-methyls.
On the left hand side, base pairings between yeast rRNA and
functionally confirmed (snR13, U24) or predicted (U18, snR52) methyl
guide snoRNAs are depicted. Spacing between the D or D' box and rRNA
sequence in each case presumably determines the location of
2'-O-methyl modification, invariably 5 bp from the end of the D/D' box
[Kiss-Laszlo et al., 1996]). If the D' box is allowed to slide one
nucleotide closer in via a single nucleotide bulge, the new placement
of the D' box could conceivably guide addition of a second 2'-O-methyl
group at the adjacent position. In each case, the single nucleotide
bulge in the snoRNA would not necessarily disrupt required base
pairings within the snoRNA/rRNA duplex.
 |



Next: Small Nucleolar RNAs in Genomes.2
Up: A Computational Screen for Yeast.2
Previous: Acknowledgments
Todd M. Lowe
2000-03-31