- ...
sequence2.1
- This chapter was co-written with Sean Eddy, and
appears in Lowe & Eddy, Nucleic Acids Research 25:
955-964, 1997.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... Yeast4.1
- This chapter
was co-written with Sean Eddy, and appears in a shortened version in
Lowe & Eddy, Science 283: 1168-1171, 1999.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
sequence4.4
- Random sequences were generated by a fifth order
Markov chain based on 6mer frequencies within the yeast genome.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... Genomes5.1
- This chapter
was co-written with Patrick Dennis as a collaboration between
the Eddy and Dennis labs. The paper appears in a shortened
version in Omer, Lowe, Russel, Ebhardt, Eddy, & Dennis,
Science (2000) (in press).
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... lab5.4
- The Dennis lab at University of British
Columbia, Department of Biochemistry and Molecular Biology, includes
Patrick Dennis, Arina Omer, Anthony Russell, and Holger Ebhart. As
collaborators on this project, the Dennis lab performed all S. acidocaldarius protein biochemistry, immunoprecipitation, and sRNA cloning.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... 5.5
- A clone containing the aFIB and aNOP56 genes
from S. solfataricus was provided by M.A. Ragan and C.W. Sensen. The Dennis lab
used Southern hybridization to identify and clone the corresponding
genes from S. acidocaldarius. The 16S rRNA sequences from S. acidocaldarius and S. solfataricus are about
90% identical. The aFIB and aNOP proteins from the two organisms are
respectively 76 and 66 percent identical. The accession numbers for
the S. acidocaldarius aFIB and aNOP genes will be obtained upon submission of the
manuscript describing this work
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... RT-PCR5.6
- The
primer AO30 (5' CTCGAGATCTGGATCCGGG 3') was 5' end-labeled with T4
polynucleotide kinase and
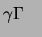 P-ATP and blocked at the 3'
end using terminal deoxynucleotidyl transferase (Gibco BRL) and dCTP.
The modified oligo was then ligated to gel purified sRNA for 16 hrs at
4�C. The ligation products were reverse transcribed with
Thermoscript RT (Gibco BRL) at 55�C for 30 min, using AO31
(5' CCCGGATCCAGATCTCGAG 3') as primer. The RNA template was
hydrolyzed with RNase H and the cDNA strand was extended with dATP
using terminal deoxynucleotidyl transferase. The extended cDNA strand
was used as template for PCR (95�C denaturation,
65�C hybridization, 72�C extension, 30 cycles)
using AO30 and AO32 [5' GCGAATTCTGCAG(T)30 3'] as primers. The
DNA products were cleaved with PstI and XhoI, ligated between the PstI
and XhoI sites of plasmid pSP72 and transformed into E. coli.
Plasmids were isolated and their sequences was determined.
P-ATP and blocked at the 3'
end using terminal deoxynucleotidyl transferase (Gibco BRL) and dCTP.
The modified oligo was then ligated to gel purified sRNA for 16 hrs at
4�C. The ligation products were reverse transcribed with
Thermoscript RT (Gibco BRL) at 55�C for 30 min, using AO31
(5' CCCGGATCCAGATCTCGAG 3') as primer. The RNA template was
hydrolyzed with RNase H and the cDNA strand was extended with dATP
using terminal deoxynucleotidyl transferase. The extended cDNA strand
was used as template for PCR (95�C denaturation,
65�C hybridization, 72�C extension, 30 cycles)
using AO30 and AO32 [5' GCGAATTCTGCAG(T)30 3'] as primers. The
DNA products were cleaved with PstI and XhoI, ligated between the PstI
and XhoI sites of plasmid pSP72 and transformed into E. coli.
Plasmids were isolated and their sequences was determined.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.