Research
Graduate
Current ResearchI have three main aspects of my research:
Common threads in my research are the kink-turn (C/D box RNAs and H/ACA box RNAs in archaea contain this motif), RNA-seq, comparative genomics, and computational prediction of RNA structures.
C/D box and H/ACA box RNAs
For both C/D box and H/ACA box RNAs, I am focused on improving computational prediction methods of these RNAs in archaea and studying their unique biology in these extremophilic species. C/D box RNAs guide 2'-O-methylation of RNA and H/ACA box RNAs guide pseudouridylation of RNA. Both of these modifications are the most common modifications in non-coding RNA, particularly rRNA and tRNAs. Functions beyond RNA modification have been discovered for these RNAs in eukaryotes, so I am exploring other possible functions for these RNAs in archaea.
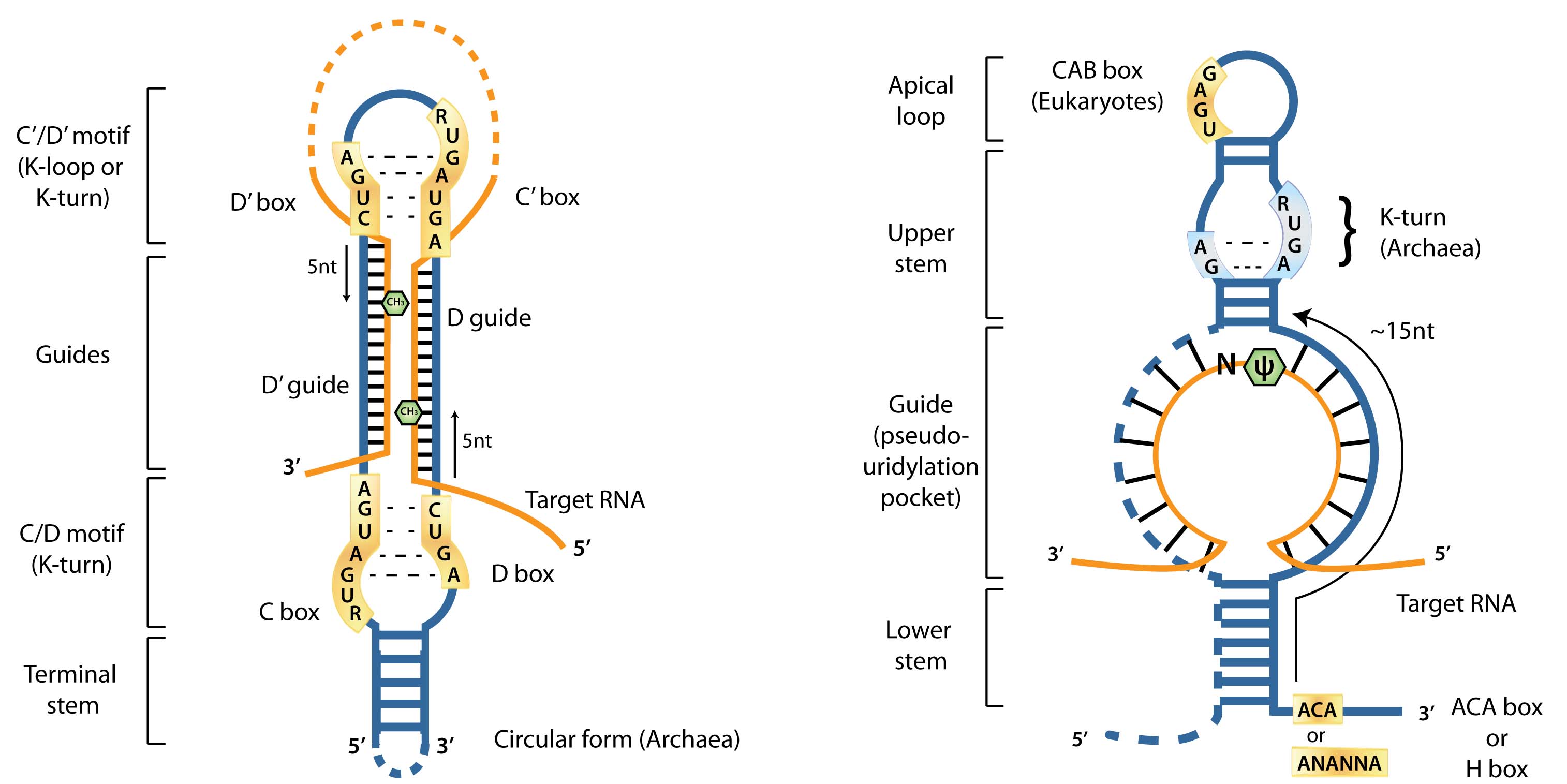
Schematic of C/D box and H/ACA box RNA features. From Lui and Lowe 2013
The RNA kink turn
The RNA kink turn is a common structural motif that is present in all three domains of life and is a key component of important ribonucleoprotein (RNP) complexes. Kink turns were first identified in the crystal structure of the Haloarcula marismortui large ribosomal subunit and are often bound by members of the L7Ae/L30 protein family. Kink turns participate in many aspects of RNA function, including translation, modification and processing of RNA and control of gene expression. David Lilley's lab at the University of Dundee has a nice page on this RNA motif.
I seek to define the diversity kink-turn motif and identify different RNP complexes where the kink turn is essential. Despite the ubiquity of kink turns and the importance of the motif in various evolutionarily conserved processes, a comprehensive study of kink turns in a transcriptome has yet to be completed. I am designing a computational search method and using a combination of co-immunoprecipitation and high throughput sequencing methods to survey sequence diversity of the kink turn motif and discover new types of kink turns. Results from the high throughput sequencing will simultaneously confirm computational predictions and provide data to refine the computational method. The recent discovery of kink turns in archaeal forms of RNase P (Cho et al 2010, PNAS) and in a new RNA family that appears to function as H/ACA box RNAs in the Pyrobaculum genus (Bernick et al, 2012, RNA) highlights the potential for new discovery of kink turns in RNA and the need for a readily available computational search tool for kink turns.
Picture of Thermococcus kodakaraensis, an archaeal species, grown in my lab.
RotationsSpring 2008 I rotated in Susan Strome's lab investigating the expression of MES-4, a chromatin regulator in Caenorhabditis elegans using immunofluorescence microscopy.
Winter 2008 I worked with David Haussler and the development of tools for the UCSC Cancer Genomics Browser.
Fall 2007 I investigated the electrostatic surface properties of DNA/RNA binding proteins with Dr. Dietlind Gerloff. Specifically, I studied families of protein domains that interact with DNA, and to which degree the electrostatic surface properties near the interaction sites are conserved.
Undergraduate
As an undergraduate I mostly conducted mathematical biology research, but I also spent a summer at UC Santa Cruz my sophomore year doing bioinformatics as part of the UC LEADS program.
Benham LabMy mentor for most of my undergraduate work was Craig Benham. My research was funded by the UC LEADS program and the NSF program VIGRE for mathematics research. Benham's research group has developed statistical mechanical methods to computationally analyze the occurrence of structural transitions in stressed DNA molecules. Stress induced DNA duplex destabilization (SIDD), the destabilization of the DNA helix from the torsional stresses caused by the superhelicity of DNA, has been found to play roles in gene regulation. I compared the SIDD profiles of orthologs from different organisms. Below is an example of a SIDD profile.
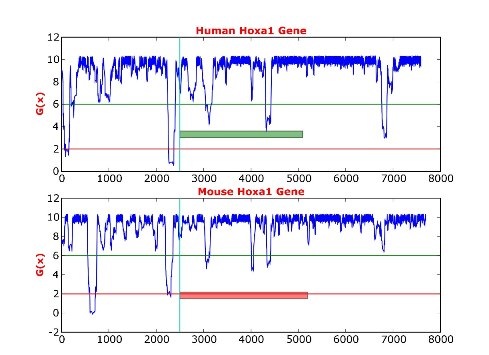
SIDD calculations quantify the topological effects on the energetics of the DNA molecule. The torsional stresses resulting from the superhelicity of DNA affect the stability of the molecule, causing the duplex to be more difficult or more easily opened at different positions. The G(x) is the free energy required to guarantee strand separation at a specific position. The lower the free energy, G(x), at a position the easier it is to open the strands.
CLIMBDuring my Senior year I was part of the NSF funded CLIMB program (Collaborative Learning at the Interface of Mathematics and Biology). It is an undergraduate research program where participants work together on projects while learning how to solve biological questions with mathematics and computational techniques. During Fall quarter, I collaborated with my peers in the program to solve problems in the areas of behavioral ecology, fisheries ecology, biofluids, cell biology, and neuroscience. During the next two quarters, my cohort and I developed our summer project on the population dynamics of vernal pool plant populations. We discussed papers, contacted other scientists studying vernal pools, and tried applying different ecological theories. During the summer we implemented the project by developing a mathematical model of the plant population and wrote a simulation to add stochastic weather effects.




