
hyper.overlap <- function(gold.matrix, cluster.assign)
{
clusters <- unique(cluster.assign)
num.clusters <- length(clusters)
num.gold <- dim(gold.matrix)[2]
pvals <- matrix(nrow=num.gold, ncol=num.clusters, data=0)
for(g in 1:num.gold)
{
gold.genes <- which(gold.matrix[,g]!=0)
for(c in 1:num.clusters)
{
cluster.genes <- which(cluster.assign == c)
overlap <- length(intersect(gold.genes, cluster.genes))
num.genes <- length(cluster.assign)
white.balls <- length(gold.genes)
black.balls <- num.genes - white.balls
k <- length(cluster.genes)
pvals[g,c] = phyper(q=overlap-1, m=white.balls, n=black.balls, k=k,
lower.tail=FALSE, log.p = TRUE) / log(10)
}
}
return(pvals)
}
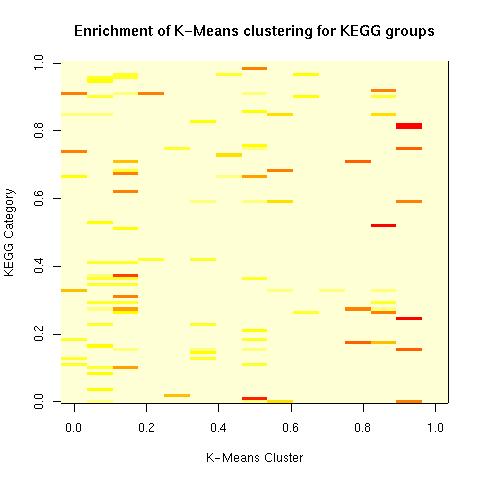
There isn't much of a one-to-one correspondence between the K-Means clusters and KEGG categories. Many KEGG categories that hit
